Most up-to-date content designed in collaboration with recognised cancer experts
Myeloproliferative neoplasms (MPNs) are a group of diseases that affect normal blood cell production in the bone marrow resulting in overproduction of one or more cell types (i.e. red cells, white cells or platelets). There are numerous different sub-types of MPNs that are distinguished from each other by the type of cell which is most affected and the genetic profile. The SureSeq Myeloid Panel targets selected key genes known to contain driver mutations for a range of MPNs including polycythaemia vera (PV), essential thrombocythaemia (ET) and myelofibrosis (MF). The gene content has been defined with input from recognised cancer experts including Professor Mike Griffiths (West Midlands
Regional Genetics Laboratory, UK) and Professor Nick Cross (National Genetics Reference Laboratory – Wessex, UK) (Table 1). To obtain the optimal sensitivity whilst maximising throughput, we have targeted hot exons where clinically relevant mutations are known and every exon for tumour suppressor, hereditary and highly implicated research-related genes. This allows for detection of previously characterised as well as novel variants in myeloid samples.
| ASXL1 |
EGLN1 |
IDH2 |
NRAS |
SRSF2 |
| CBL |
EPAS1 |
JAK2 |
RUNX1 |
TET2 |
| CALR |
EPOR |
KIT |
SETBP1 |
TP53 |
| CSF3R |
EZH2 |
KRAS |
SF3B1 |
U2AF1 |
| DNMT3A |
IDH1 |
MPL |
SH2B3 |
VHL |
Table 1: The SureSeq Myeloid Panel targets 25 genes implicated in a variety of MPNs.
Time and cost saving solution
Genetic analysis of single nucleotide variants (SNVs) and indels in myeloid-implicated genes is a reliable way of distinguishing between different subtypes of MPNs. Instead of assaying for single genes in a sequential manner, the mutational status of 25 genes can be rapidly and simultaneously determined with the use of the SureSeq Myeloid Panel. This approach therefore offers both a time and cost-effective solution.
Sensitive and reproducible variant detection even in heterogeneous samples
Heterogeneous cancer samples pose significant challenges as alleles are likely to be present at a lower fraction than what would be expected for standard germline variants. Samples typically contain a mixture of cancer and normal cells; moreover, cancer can consist of several molecularly distinct clones. In order to detect alleles that contribute only a small percentage to the reads at any locus, a highly uniform and sensitive enrichment is required. The SureSeq Myeloid Panel has been validated with samples from the National Institute for Biological Standards and Control (NIBSC) and has been shown to accurately detect alleles down to 1% minor allele fraction (MAF) at a read depth of >1000x (Table 2).
| NIBSC JAK2 V617F Sample |
% variant detected |
Wild type reads |
Variant reads |
95% confidence intervals |
| 10% |
13.6 |
1443 |
227 |
12 - 15.3 |
| 5% |
7.1 |
1180 |
90 |
5.8 - 8.6 |
| 1% |
1.6 |
1504 |
24 |
1.1 - 2.3 |
| 0.10% |
0.6 |
1380 |
8 |
0.3 - 1.1 |
| 0% |
0.0 |
1630 |
0 |
0.0 - 0.2 |
Table 2: Data generated from 24 samples run on an Illumina MiSeq®. The SureSeq Myeloid Panel allowed the detection of alleles at 1% sensitivity with high confidence (95% confidence interval 1.1 – 2.3%).
A typical laboratory workflow might be to pool and run 24 samples in a single Illumina MiSeq lane. The SureSeq Myeloid Panel can reliably and routinely detect somatic mutations down to 1% MAF under these conditions at the following sites of interest in MPNs:
- JAK2 (exon 12 – AAs 536-547)
- JAK2 (V617F)
- MPL (W515)
- CALR (exon 9 indels)
- KIT (D816V)
- TET2 (R550)
Analysis in clinical research samples has shown that the panel is able to reliably detect not only SNVs but also deletions of up to 52 bp, which are particularly informative in the CALR gene (Table 3).
| Sample |
Known mutation |
Mean target coverage |
% MAF detected |
| Sample 1 |
52 bp del in CALR exon 9 |
2351 |
23% |
| Sample 2 |
52 bp del in CALR exon 9 |
1315 |
8.3% |
| Sample 3 |
JAK2 V617F |
1500 |
34% |
| Sample 4 |
JAK2 V617F |
1254 |
11% |
Table 3: The SureSeq Myeloid Panel generated data concordant with previous findings. Data obtained using the panel on clinical research samples containing known CALR and JAK2 mutations. 24 samples were run on a MiSeq lane using 500 ng input DNA. Samples provided by the National Genetics Reference Laboratory – Wessex, UK.
Fast and easy workflow
Hybridisation-based enrichment is now well recognised as providing superior results over amplicon-based enrichment technology. To date, the protocol has required more DNA and the library preparation procedure has been longer and more complex. In combination with the OGT SureSeq Library Preparation Kit, these issues have been addressed. There are fewer hands-on steps, turnaround times have been significantly improved, and the panel has been optimised to work with as little as 500 ng of genomic DNA derived from whole blood.
SureSeq Interpret Software — OGT’s powerful, standalone data analysis package — is provided free with the SureSeq Myeloid Panel and allows the conversion of FASTQ files into an intuitive interactive report. The user-friendly report makes it easy to set up complex filtering rules with multiple parameters.
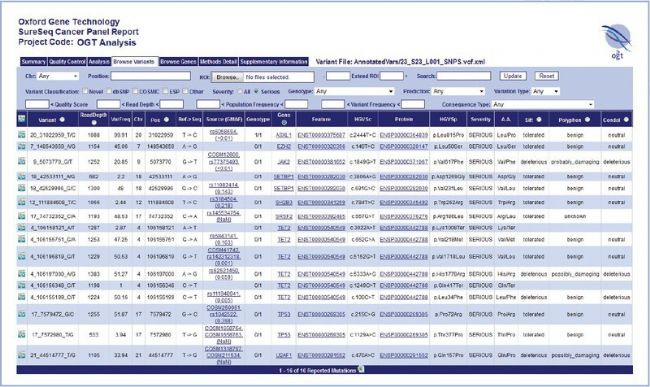
Figure 1: The SureSeq Interpret Report enables simple and rapid identification of meaningful results.
Variants are fully annotated with links to various databases (e.g. dbSNP, COSMIC, Genecards and OMIM) providing results in context. Each variant can be reviewed in the Integrative Genomics Viewer (IGV) from the Broad Institute (Figure 2).
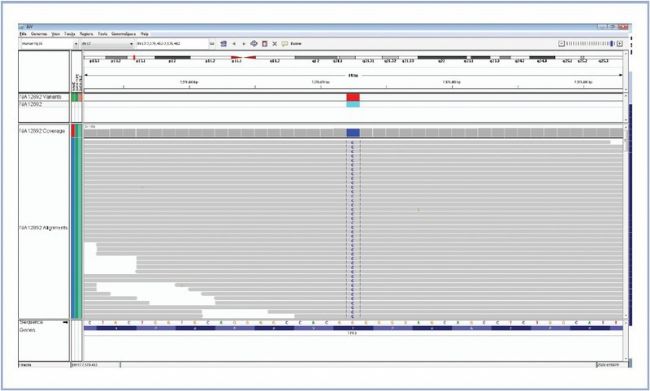
Figure 2: Fast visual confirmation of variants using IGV from the Broad Institute.
Excellent coverage uniformity
Enrichment assay optimisation is a crucial step in ensuring accuracy and sensitivity of targeted sequencing. Where regions are poorly enriched, they will generate fewer sequencing reads. If a variant falls into a region that is not covered at all, or covered by only a few reads, that variant is likely to be missed. OGT’s expert bait design ensures efficient and more uniform capture of all targeted regions, so that all variants present can be called with maximum confidence. This has been demonstrated on the CALR gene, which is commonly mutated in various MPNs. It is critical to identify key CALR indels (types 1 & 2 causing a frameshift) as well as increasingly recognised point mutations in this gene. The SureSeq Myeloid Panel delivers superior performance to panels designed using standard algorithms by ensuring uniform coverage over the regions of interest (Figure 3).
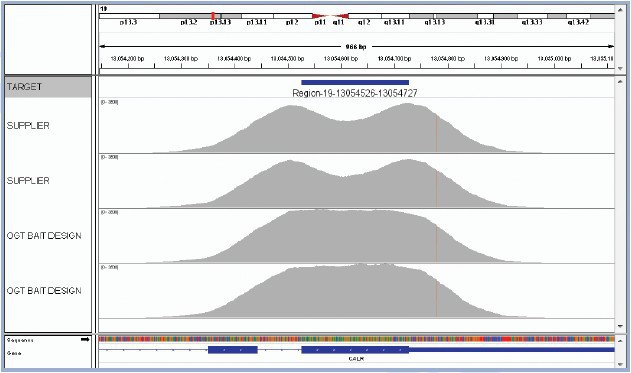
Figure 3: OGT’s expert bait design delivers improved uniformity of coverage. The image shows exon 9 of CALR. The top two captures have been completed using baits designed with standard commercially available software. They have a considerable dip in coverage in the middle of the exon due to the fact it presents a low complexity region with low nucleotide diversity. Most algorithms would want to avoid such regions in the design. However, OGT’s superior bait design can increase the evenness of coverage of such regions.
bio-equip.cn







