Array content fully optimised and research-validated
CytoSure disease-focused research arrays have been designed and optimised in collaboration with leading molecular genetics experts at Emory University. Each gene is targeted by multiple exon-specific probes, allowing accurate detection of copy number variations (CNVs) encompassing single and multiple exons. While Sanger sequencing, and increasingly targeted next generation sequencing (NGS), are being used to detect specific point mutations, it is not possible to use these techniques to accurately detect CNV. As such, many molecular genetics researchers are choosing to complement sequencing studies with array comparative genomic hybridisation (aCGH) — the gold-standard for CNV detection.
CytoSure arrays utilise 60mer oligonucleotide probes, which have been shown to offer higher signal-to-noise ratios through increased specificity and sensitivity1. To further improve performance, each array has been verified by Emory to ensure only the best performing probes were used.
The optimised array design coupled with the powerful CytoSure Interpret Software that accompanies each array ensures sensitive detection of aberrations as small as a few hundred bases within each gene exon.
These arrays provide a cost-effective, routine option for detailed research into small, exon-specific aberrations.
"OGT has the experience and expertise required to optimise the design of microarrays, which is why we chose to partner with them". Dr Madhuri Hegde, Executive Director, Emory Genetics Lab, United States of America
Highly targeted optimised probes
Using a proprietary probe design algorithm, it is possible to design highly targeted, optimised probes throughout the majority of the genome. A range of probe performance metrics were evaluated to ensure optimal array performance. A poorly performing probe can result in inaccurate data and even false calls. All of our probes are first tested in silico and scored on quality. Probes with the highest score are printed on an array and tested in the laboratory. These probes are ranked on performance, and only the most accurate, best performing probes are used in the final designs. Probes have been designed to target over 128,000 exons across 4645 genes (the complete probe set is found on the Medical Research Exome Array) and a subset of these probes are then used for the disease-focused research arrays. Using optimised probes enables the detection of small amplifications and deletions.
The targeted, optimised probes deliver:
An average resolution of 1 probe per 125bp in targeted exons.
A minimum of 4 probes per targeted exon
Targeting of exome flanking regions (150bp 5’ and 3’ of each exon)
Targeting of introns, and gene flanking regions (average resolution of 1 probe per 1kb)
Coverage of genomic backbone
Easy data interpretation
CytoSure Interpret Software is a powerful, easy-to-use package for the analysis of aCGH data. Innovative features such as the Accelerate Workflow enable standardised and automated data analysis, including automatic aberration detection and classification. It includes extensive annotation tracks covering syndromes, genes, exons, CNVs and recombination hotspots — each of which link to publicly available databases such as ISCA, Decipher and the Database of Genomic Variants (Figure 1) providing results in context. It is possible to select which tracks are displayed allowing only tracks of specific interest to be viewed (e.g. syndrome-specific tracks) ensuring easy data interpretation. Each track can reference NCBI36 (hg18), GRCh37 (hg19) or GRCh38 (hg38) information. Annotations within a track can be coloured allowing easy visualisation. It is also possible to customise which information from the tracks is saved in the report (e.g. how many common variants overlap with an aberration).
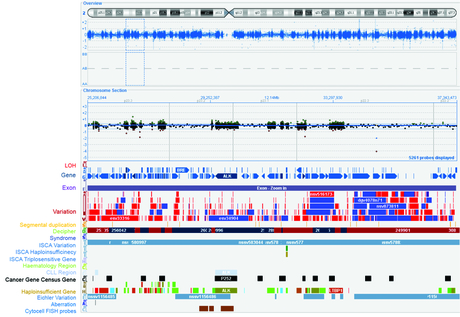
Figure 1: Chromosome overviews are clearly displayed in CytoSure Interpret Software. Shown here is an overview of the distribution of probes on chromosome 2 in the CytoSure Medical Research Exome array and the currently available standard tracks. These fully customisable tracks simplify the interpretation of aberrations.
The complete solution
All CytoSure arrays have been validated using CytoSure Genomic DNA Labelling Kits; these labelling kits have been uniquely developed and optimised to enable rapid delivery of high-quality results with excellent signal-to-noise ratios. Two formats are available; the CytoSure Genomic DNA Labelling Kit is sufficient for 24 samples and is ideal for labs running one or two arrays a week. For high-throughput labs, the CytoSure HT Genomic DNA Labelling Kit is recommended as its plate-based protocol allows simultaneous labelling of 96 samples. To achieve the best quality data possible, it is recommended that CytoSure arrays are used in conjunction with CytoSure Genomic DNA Labelling Kits.
bio-equip.cn





