Amplification technology developed by IDT that facilitates SNP genotyping applications, high levels of multiplexed amplification, and detection of rare alleles.
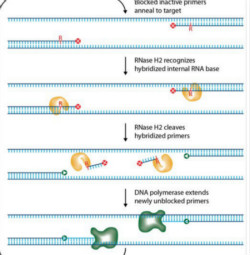
RNase H-dependent PCR (rhPCR) relies on cleavage of a 3’-blocked primer by RNase H2 to effect primer activation before target amplification. The cleaved, unblocked primer is then available to support extension and replication in the polymerase chain reaction (PCR). Cleavage occurs at the 5’ end of a single RNA base that is incorporated near the 3’ end of the primer, leaving a 3’-hydroxyl on the terminal DNA base. A blocking moiety included downstream of the RNA base can block either direct extension of the primer or replication by the polymerase during subsequent cycles of PCR. The blocked primer can only be cleaved when bound to its complementary target sequence and at elevated temperature, which strongly reduces the formation of primer-dimers or misprimed species that can result in unwanted amplification products.
Blocked-cleavable rhPCR primers (rhPrimers) and the thermostable RNase H2 enzyme required for rhPCR are available from IDT.
High specificity is often achieved with the polymerase chain reaction (PCR); however, sometimes it is necessary to position primers at suboptimal locations in the target. This can result in the formation of primer dimers and/or undesired amplification of homologous sequences.
IDT scientists have developed RNase H2–dependent PCR, a method for increasing PCR specificity and eliminating primer dimers by using RNase H2 from Pyrococcus abyssi (P.a.) and blocked primers that contain a single ribonucleotide residue. The blocked primers are activated when cleaved by the RNase H2 enzyme. Cleavage occurs at the 5’ end of the RNA base after primer hybridization to the target DNA. Because the primers must hybridize to the target sequence before they are cleaved, they are unable to form primer dimers and the requirement for high target complementarity reduces amplification of closely related sequences (Figure 1). rhPCR is more sensitive than allele-specific PCR for detection of single-nucleotide polymorphisms (SNPs). Superior allele discrimination is achieved when the mismatched base is positioned at the RNA:DNA base pair [1].
Pyrococcus abyssi is an extreme thermophile, so the P.a. RNase H2 enzyme has optimal activity in range of 70–75°C and is functional in rhPCR between 50°C and 75°C. P.a. RNase H2 has very low activity at room temperature (~1000X less active). Therefore, use of this enzyme to perform primer activation confers a "hot start" character to the PCR. P.a. RNase H2 is functional in most PCR buffers and can be added directly to the PCR master mix. The enzyme functions in real time, making the method a transparent change to standard PCR with primer cleavage occurring in the background during each anneal/extend cycle. [1].
Figure 1. Schematic Representation of rhPCR
References
-
Dobosy JR, Rose SD, Beltz KR, Rupp SM, Powers KM, Behlke MA, and Walder JA (2011)RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnol, 11:80.
bio-equip.cn